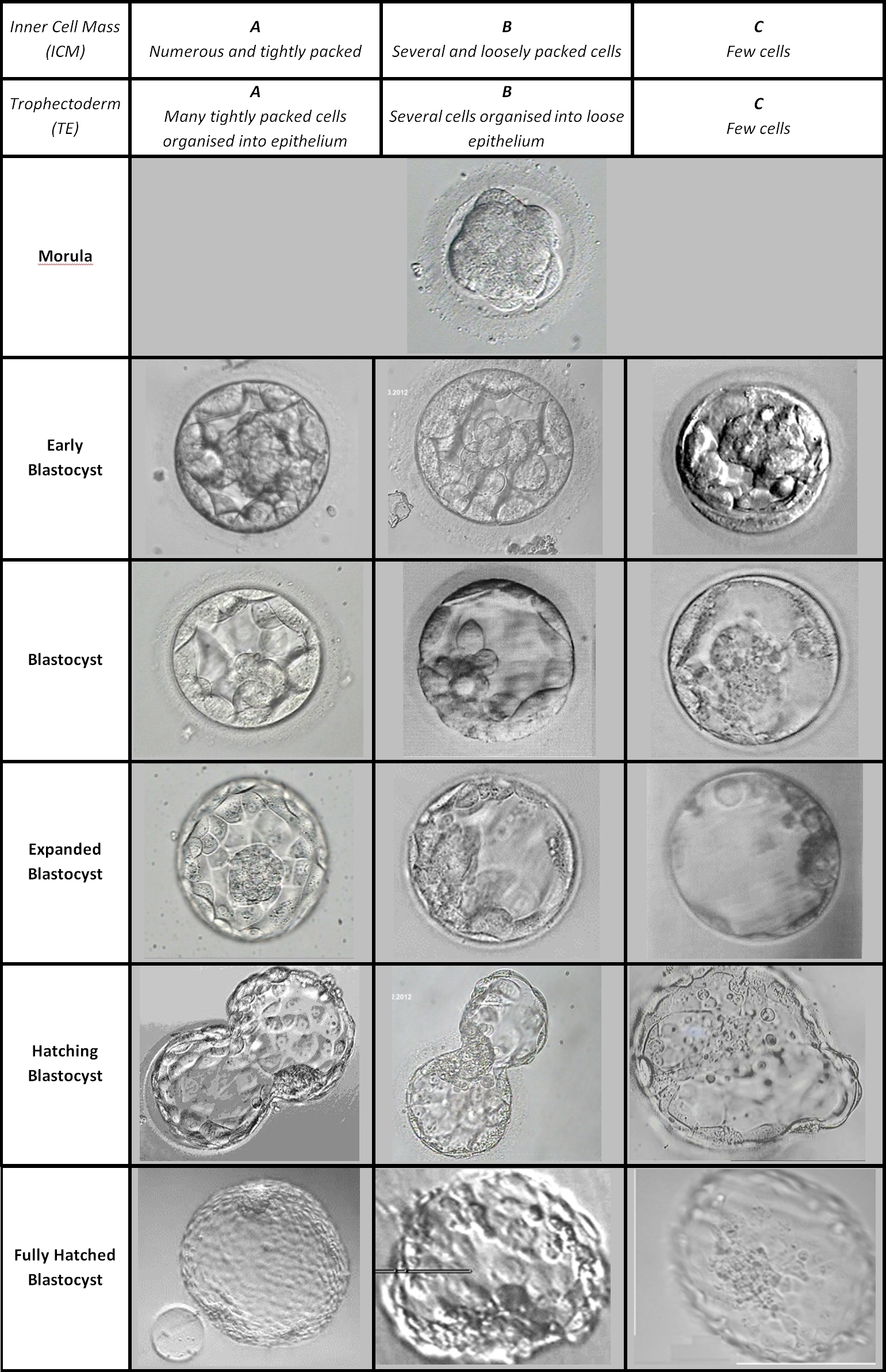
A lot of people are interested in embryo selection. Why? When people reproduce through IVF, they will generate more than one viable embryo. Which one to choose? The general approach is two-fold: phenotypical criteria and genetic. Phenotypic criteria are typified by the embryo grading scale:
So you look at the embryo in a microscope and a clinician (embryologist) makes some judgment calls as regards the above 3 criteria. This then results in an overall grade, where you want the top scoring embryo. In practice, you begin with the best one, and if it fails to plant, then you try the 2nd best one. Repeat until you succeed (live birth) or run out of eggs. If you run out of eggs, you can harvest some more unless you are too old.
Genetic selection involves taking a small sample of cells from the embryo, then reading the DNA and making some predictions about what the child is likely to be like. This idea used to be sci-fi but has been used for complex types — usually the most important ones — for half a decade already in humans, and much longer in animals and plants (just search for “genomic selection”). This allows you to select against heritable diseases, as well as for or against psychological (intelligence, mental health) and physical traits (height, obesity). You can find a lot of academics being oh so concerned about this technology and saying it’s not currently very good, while parents are lining up to get the best child they can. A lot of countries semi-ban the technology, but one can simply fly to a country where it is legal (USA, Asia), or have the company “do the selection” there. The future of humanity depends on these biotechnologies, so it is an important battle.
This tissue sampling procedure does some level of harm to the embryo, but many people are willing to take the trade-off in order to get precious predictions. With cutting edge sequencing technology, probably a single cell is sufficient, but the routine practice is to take about 5 cells.
The problem with embryo selection is that the primary limiting factor is the number of eggs. Every woman is born with her eggs, and they decay with age. By age 40, it can be difficult to get pregnant due to the poor quality of the remaining eggs. At 50, it is nearly impossible without IVF and maybe with it as well.
So in terms of embryo selection, we desperately need more eggs. That’s why there are so many companies working on trying to make egg cells from other cells. When this succeeds, we can generate a large number of embryos, grow them all, biopsy, read the genetic data, and forecast. This gametogenesis — generation of gametes (sex cells, sperms and eggs) from other cells — is probably some 5-10 years away from clinical practice. You may have read that it has been done already in various animals, even same-sex ones, but the technology is somewhat different for humans than the rodents. It is also difficult to get through all the red tape blocking this work. For these reasons, I expect it will take a number of years before this technology is available for clinical use.
But there is another way to improve the process currently. Let’s first make a list of all the factors that embryo selection’s efficiency for improving complex traits depends on:
- Number of eggs
- Quality of eggs
- Number of sperm
- Quality of sperm
- Accuracy of genetic reading
- Accuracy of genetic prediction model
- IVF procedures (freezing/thawing, implantation rate, growing the embryo etc.)
We currently cannot do that much about (1-2). What about (3)? We have literally millions of sperm, so that’s not really much of a limiting factor. However, (4) is. Men produce a lot of sperm, and much of them are of low quality. The way evolution has dealt with this is to design women’s uterus to be kind of hostile to sperm. This hostile environment then selects for the more fit sperm, which are the ones that can swim far enough in this environment to reach the egg and enter it.
For many people doing IVF, the reason is that the man’s sperm is too low quality to achieve natural pregnancy. It’s estimated that some 25-50% of infertility is due to male factors. For this reason, a method of directly putting the sperm inside the egg was developed (ICSI).
Still, considering that we have so many sperm, it seems wise to pick good sperm to put into our few very precious eggs. So how can we mimic, and ideally improve upon, the natural selection that happens in the female body? There’s actually quite a few methods for doing this. Here’s two recent review papers for those interested in the details:
- Vaughan, D. A., & Sakkas, D. (2019). Sperm selection methods in the 21st century. Biology of reproduction, 101(6), 1076-1082.
Natural sperm selection in humans is a rigorous process resulting in the highest quality sperm reaching, and having an opportunity to fertilize, the oocyte. Relative to other mammalian species, the human ejaculate consists of a heterogeneous pool of sperm, varying in characteristics such as shape, size, and motility. Semen preparation in assisted reproductive technologies (ART) has long been performed using either a simple swim-up method or density gradients. Both methodologies provide highly motile sperm populations; however neither replicates the complex selection processes seen in nature. A number of methods have now been developed to mimic some of the natural selection processes that exist in the female reproductive tract. These methods attempt to select a better individual, or population of, spermatozoa when compared to classical methods of preparation. Of the approaches already tested, platforms based upon sperm membrane markers, such as hyaluronan or annexin V, have been used to either select or deselect sperm with varied success. One technology that utilizes the size, motility, and other characteristics of sperm to improve both semen analysis and sperm selection is microfluidics. Here, we sought to review the efficacy of both available and emerging techniques that aim to improve the quality of the sperm pool available for use in ART.
- Baldini, D., Ferri, D., Baldini, G. M., Lot, D., Catino, A., Vizziello, D., & Vizziello, G. (2021). Sperm Selection for ICSI: Do We Have a Winner?. Cells, 10(12), 3566.
In assisted reproductive technology (ART), the aim of sperm cells’ preparation is to select competent spermatozoa with the highest fertilization potential and in this context, the intracytoplasmic sperm injection (ICSI) represents the most applied technique for fertilization. This makes the process of identifying the perfect spermatozoa extremely important. A number of methods have now been developed to mimic some of the natural selection processes that exist in the female reproductive tract. Although many studies have been conducted to identify the election technique, many doubts and disagreements still remain. In this review, we will discuss all the sperm cell selection techniques currently available for ICSI, starting from the most basic methodologies and continuing with those techniques suitable for sperm cells with reduced motility. Furthermore, different techniques that exploit some sperm membrane characteristics and the most advanced strategy for sperm selection based on microfluidics, will be examined. Finally, a new sperm selection method based on a micro swim-up directly on the ICSI dish will be analyzed. Eventually, advantages and disadvantages of each technique will be debated, trying to draw reasonable conclusions on their efficacy in order to establish the gold standard method.
These are some of the methods:
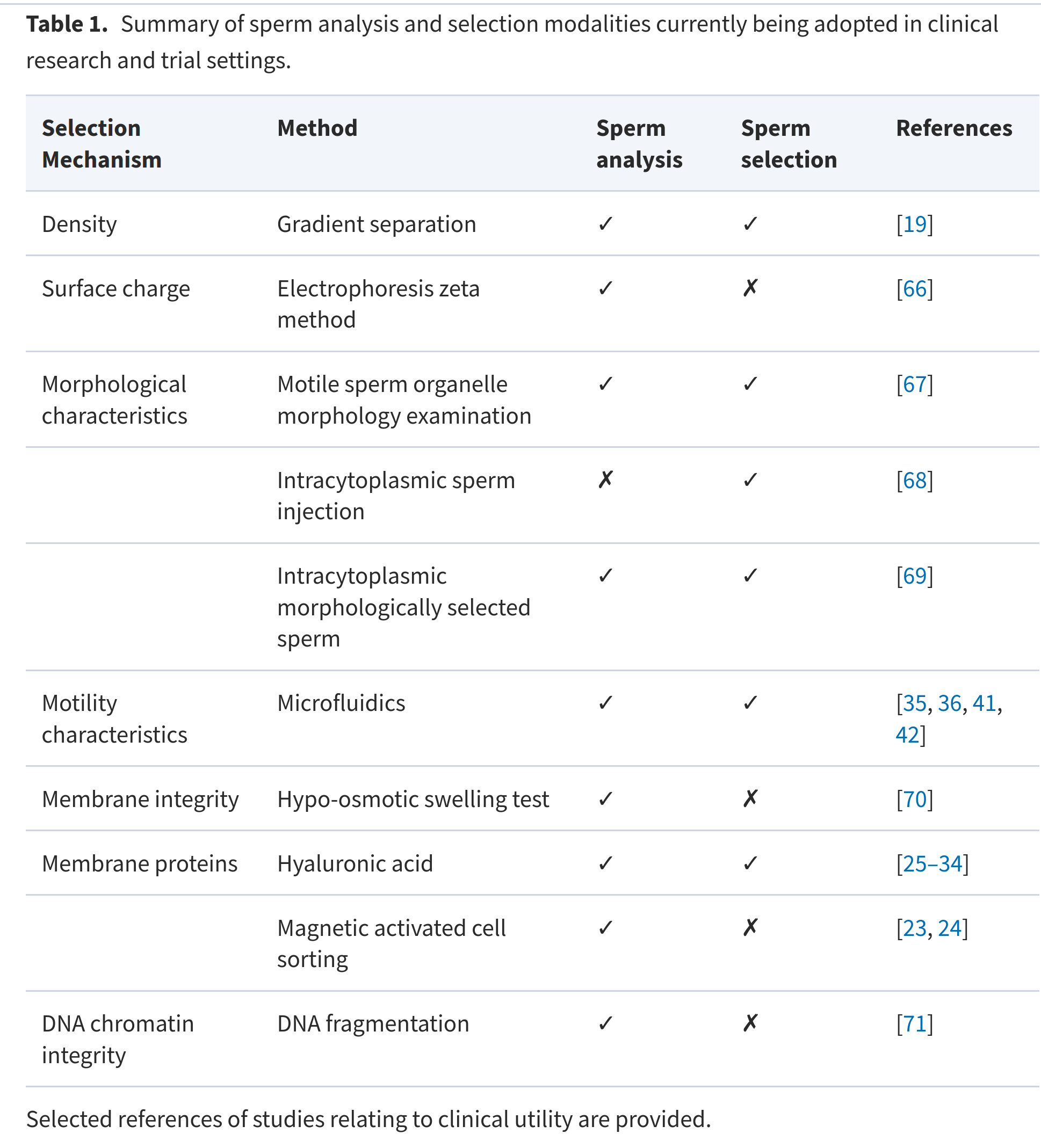
They range from simple swimming tests to various chemical and physical methods of assessing the vitality, swimming ability, DNA integrity of the sperm. Here’s the swimming one:

The various authors argue about the best methods. Science as it should be. But it is also clear that none of these are particularly impressive. It is one thing is to not waste an egg on a sperm that will result in a non-viable embryo, another thing is to pick a sperm with good genetics for diseases, physical, and psychological traits. Can it be done? As far as I can tell, none of the chemical methods listed above have tried to get at this. However, it is theoretically possible. How? One can sequence sperm (single cells) and thus assess their genetics, and one can also take a lot of high-resolution photos/videos of sperm. As such, it is also possible to build a machine learning model to train from how the sperm looks and behaves into its underlying genomics. Here’s a figure from a 2021 study.
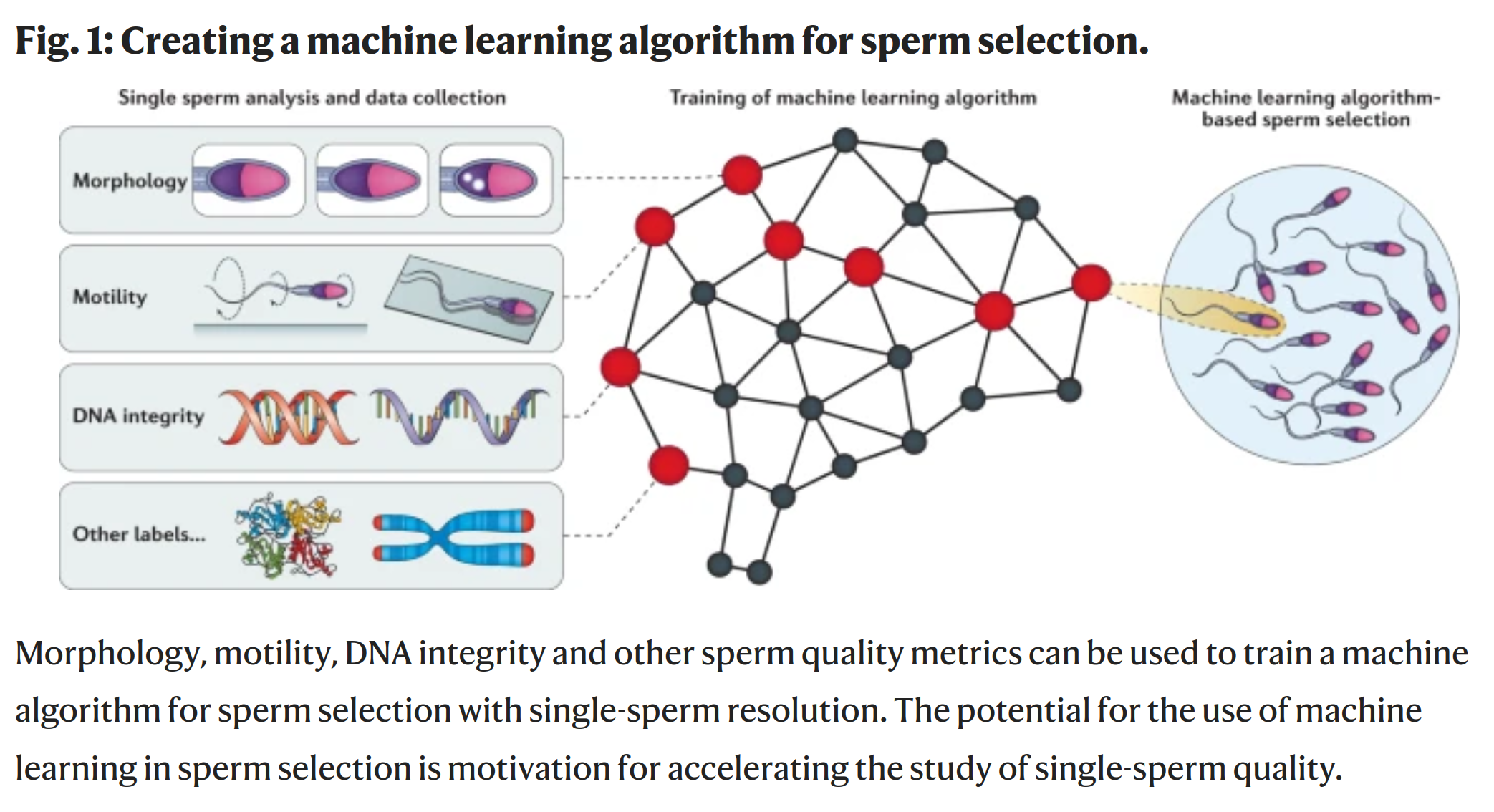
The potential is enormous. If we can track and score 1000 sperm with a weakly predictive model, this can result in large gains. Consider a model that has an accuracy of predicting the polygenic score (PGS) for height from sperm image data with r = .10. It’s pretty terrible but it’s a slight signal. But if we have 1000 sperm, and we pick the top one, then on average we will pick a sperm that’s 3.24 standard deviations (z) above the mean. Thus, the expected gain will be the accuracy of the PGS times the accuracy of the sperm-ML model. So if we assume the height model’s accuracy is .55 (2020 study), the expected gain is .10 * .55 * 3.24 = 0.18 z. Not exactly nothing, but useful. The sperm tracking technology is probably fairly cheap, so we could increase the sperm selection to best of 10,000, and this would give us: .10 * .55 * 3.85 = 0.21 z. Thus, there are large diminishing returns to increasing the sperm count for selection. The main improvement will come from improving the sperm-model as well as the PGS for the phenotype in question (the intelligence one is about .30, so the IQ gain would be about 1.7 IQ for best of 1000 sperm). It is currently unknown how good such a machine learning model based on sperm image data could become, but we should find out. This 2008 paper already showed that one can use proteins to figure out which sperm came from high vs. low fertility bulls, so I don’t see why this could not also be done for other complex traits based on image data.
Can we do genetic selection for sperm directly?
But there is another unexplored method. In the above, we tried to select sperm only on phenotype, if only as a proxy for the genotype. Is it possible to select directly on the genotype? It seems not. When you sequence a sperm, you kill it, so if you found it out had a killer +3 z for intelligence, you just killed it, so you can’t use it, and that’s a bit of a bummer. However, there is a way. Because gamete only have 1 copy of each chromosome, it means that they are generated in pairs. Here’s how it looks like:
In egg generation (oogenesis), you begin with a proto-egg of sorts (oogonium). Then you make the primary oocyte, which still has 2 copies of each chromosome (diploid). Then to create the ootid, recombination happens, hence the mixed chromosome of different colors. Notice the small cells being ejected called polar bodies. These have the complementary DNA of the egg. For instance, if on chromosome 1, the egg’s first half comes from the mother’s chromosome 1a, then the polar body will have the genetic material from the mother’s chromosome 1b there. In other words, by analogy with twin particles in quantum mechanics, if you sequence the polar body, and you know the mother’s genome, you can infer what the DNA of the egg must be (e.g., if you know the mother has AT at a given loci, and you observe the polar body has T, you know the egg has A). In fact, many researchers have already sequenced polar bodies. For embryo selection, this method is not very useful. You only have the few eggs you have to work with, so picking the best egg among them is not of much use. You will use all of them to generate embryos anyway. However, for sperm, this is another matter. If you have 1000 sperm, and you can read their DNA, you can pick the top one to use on your precious embryos. Again, recall that if this can be done, every embryo starts with a half genome that’s some +3 z for whatever polygenic trait(s) you are interested in. That’s a big deal!
The problem from a practical perspective is that the complimentary brother sperms are hard to find in a pool of 200 million sperm. I see two approaches. First, we use the same idea as above. We get some sperm, record them, then sequence them all. Using the DNA, we infer which ones are complementary brothers, and then train a machine learning model to find these pairs in the wild. The problem here, I think, is that when men ejaculate some 200 million sperm, it is too difficult to measure all of these to be able to do an exhaustive search. If the complementary brother sperm are not found in the sample of say 1000 sperm we analyze, it will not be possible to find them.
Second, if we could monitor the generation of sperm in vitro (in a lab), we could simply see which ones are generated in pairs, and then compartmentalize them. Once the complementary brother sperm are compartmentalized, we sequence one of them, and infer the genotype of the remaining one. As far as I can tell (search1, search2), no one is pursuing this idea, but the potential for the improvement of embryo selection is huge. I imagine one could do this by taking a biopsy from men’s testicles to get the spermatogonium cells (refer to figure above), and then monitor the process in a lab. It would probably not be pleasant, but if women have to undergo painful hormone stimulation and egg retrieval to get better or any kids, then men should also be willing to take a needle in their balls.