Neuroscience enjoys a good public perception of scientific rigor. Unfortunately, it’s undeserved. Statistically speaking, a scientific field cannot be rigorous when it is underpowered and has a high researcher degree of freedom. This combo is exactly what neuroscience has, so we get stuff like:
-
Button, K. S., Ioannidis, J. P., Mokrysz, C., Nosek, B. A., Flint, J., Robinson, E. S., & Munafò, M. R. (2013). Power failure: why small sample size undermines the reliability of neuroscience. Nature reviews neuroscience, 14(5), 365-376.
A study with low statistical power has a reduced chance of detecting a true effect, but it is less well appreciated that low power also reduces the likelihood that a statistically significant result reflects a true effect. Here, we show that the average statistical power of studies in the neurosciences is very low. The consequences of this include overestimates of effect size and low reproducibility of results. There are also ethical dimensions to this problem, as unreliable research is inefficient and wasteful. Improving reproducibility in neuroscience is a key priority and requires attention to well-established but often ignored methodological principles.
- Empirically, we estimate the median statistical power of studies in the neurosciences is between ∼8% and ∼31%.
To expose the researcher degrees of freedom, one can give many teams the same dataset and same research questions and tell them to analyze the data how they see fit, as done previously for football racism. There is such a study:
-
Botvinik-Nezer, R., Holzmeister, F., Camerer, C. F., Dreber, A., Huber, J., Johannesson, M., … & Avesani, P. (2020). Variability in the analysis of a single neuroimaging dataset by many teams. Nature, 1-7.
Data analysis workflows in many scientific domains have become increasingly complex and flexible. To assess the impact of this flexibility on functional magnetic resonance imaging (fMRI) results, the same dataset was independently analyzed by 70 teams, testing nine ex-ante hypotheses. The flexibility of analytic approaches is exemplified by the fact that no two teams chose identical workflows to analyze the data. This flexibility resulted in sizeable variation in hypothesis test results, even for teams whose statistical maps were highly correlated at intermediate stages of their analysis pipeline. Variation in reported results was related to several aspects of analysis methodology. Importantly, meta-analytic approaches that aggregated information across teams yielded significant consensus in activated regions across teams. Furthermore, prediction markets of researchers in the field revealed an overestimation of the likelihood of significant findings, even by researchers with direct knowledge of the dataset. Our findings show that analytic flexibility can have substantial effects on scientific conclusions, and demonstrate factors related to variability in fMRI. The results emphasize the importance of validating and sharing complex analysis workflows, and demonstrate the need for multiple analyses of the same data. Potential approaches to mitigate issues related to analytical variability are discussed.
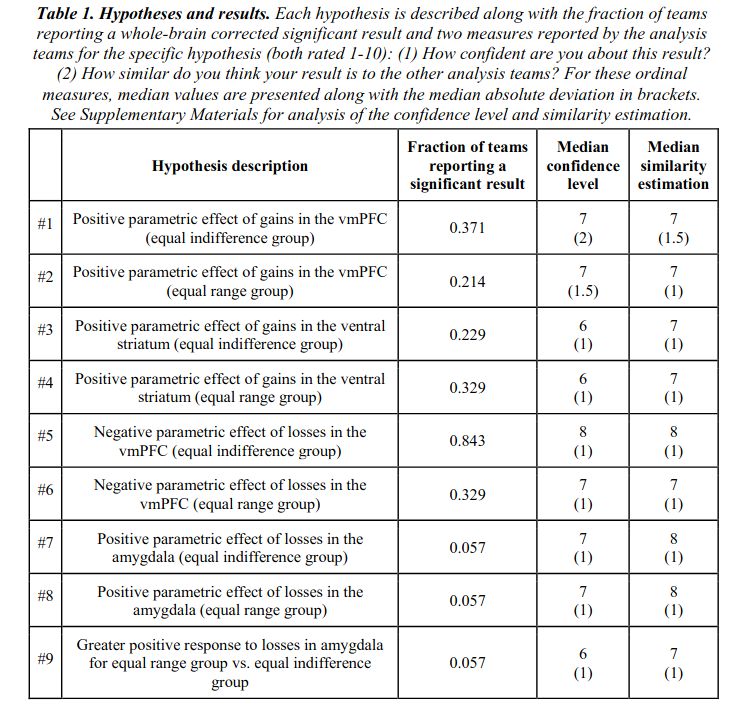
Thus, there was only 1 hypothesis that received mostly consistent support across teams, at 84% finding p < .05. Pathetic!
Previously, geneticists made some similar mistakes when they published 1000s of unreplicable candidate gene papers. The problem was they could choose which variants to test at will, and thus delude themselves. One group of researchers are advocating the same approach for neuroscience with regards to brain regions, and it ain’t pretty:
- Marek, S., Tervo-Clemmens, B., Calabro, F. J., Montez, D. F., Kay, B. P., Hatoum, A. S., Donohue, M. R., Foran, W., Miller, R. L., Feczko, E., Dominguez, O. M., Graham, A., Earl, E. A., Perrone, A., Cordova, M., Doyle, O., Moore, L. A., Conan, G., Uriarte, J., … Dosenbach, N. U. F. (2020). Towards Reproducible Brain-Wide Association Studies. BioRxiv, 2020.08.21.257758
Magnetic resonance imaging (MRI) continues to drive many important neuroscientific advances. However, progress in uncovering reproducible associations between individual differences in brain structure/function and behavioral phenotypes (e.g., cognition, mental health) may have been undermined by typical neuroimaging sample sizes (median N=25)1,2. Leveraging the Adolescent Brain Cognitive Development (ABCD) Study3 (N=11,878), we estimated the effect sizes and reproducibility of these brain wide associations studies (BWAS) as a function of sample size. The very largest, replicable brain wide associations for univariate and multivariate methods were r=0.14 and r=0.34, respectively. In smaller samples, typical for brain wide association studies, irreproducible, inflated effect sizes were ubiquitous, no matter the method (univariate, multivariate). Until sample sizes started to approach consortium levels, BWAS were underpowered and statistical errors assured. Multiple factors contribute to replication failures4,5,6; here, we show that the pairing of small brain behavioral phenotype effect sizes with sampling variability is a key element in widespread BWAS replication failure. Brain behavioral phenotype associations stabilize and become more reproducible with sample sizes of N>2,000. While investigator initiated brain behavior research continues to generate hypotheses and propel innovation, large consortia are needed to usher in a new era of reproducible human brain wide association studies.
While the above paints a very bleak picture in terms of statistical rigor, what about the metrics used? Are they even reliable? Not so, says a big study:
-
Elliott, M. L., Knodt, A. R., Ireland, D., Morris, M. L., Poulton, R., Ramrakha, S., … & Hariri, A. R. (2020). What Is the Test-Retest Reliability of Common Task-Functional MRI Measures? New Empirical Evidence and a Meta-Analysis. Psychological Science, 0956797620916786.
Identifying brain biomarkers of disease risk is a growing priority in neuroscience. The ability to identify meaningful biomarkers is limited by measurement reliability; unreliable measures are unsuitable for predicting clinical outcomes. Measuring brain activity using task functional MRI (fMRI) is a major focus of biomarker development; however, the reliability of task fMRI has not been systematically evaluated. We present converging evidence demonstrating poor reliability of task-fMRI measures. First, a meta-analysis of 90 experiments (N = 1,008) revealed poor overall reliability—mean intraclass correlation coefficient (ICC) = .397. Second, the test-retest reliabilities of activity in a priori regions of interest across 11 common fMRI tasks collected by the Human Connectome Project (N = 45) and the Dunedin Study (N = 20) were poor (ICCs = .067–.485). Collectively, these findings demonstrate that common task-fMRI measures are not currently suitable for brain biomarker discovery or for individual-differences research. We review how this state of affairs came to be and highlight avenues for improving task-fMRI reliability.
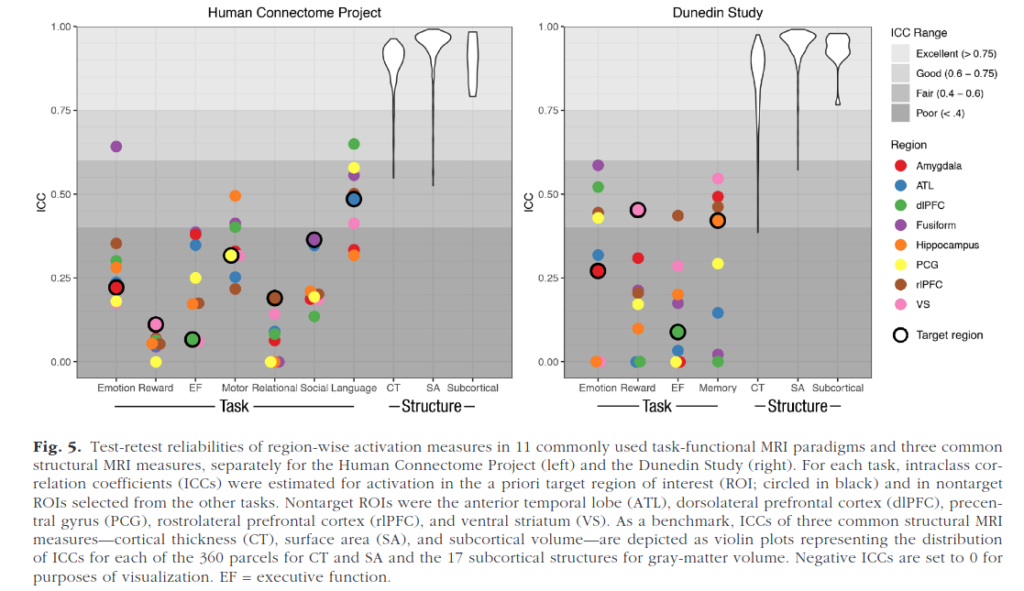
Comically poor test-retest reliability of functional MRI (fMRI), so bad it makes even psychologists laugh out loud. So when you see something based on that, best to ignore. Structural MRI (sMRI) are pretty good though, but beware of the long tail see in the figure above.
From the perspective of contrarianism, we should note that there is something that replicates beautifully in neuroscience, in large UKBB study:
-
Cox, S. R., Ritchie, S. J., Fawns-Ritchie, C., Tucker-Drob, E. M., & Deary, I. J. (2019). Structural brain imaging correlates of general intelligence in UK Biobank. Intelligence, 76, 101376.
The associations between indices of brain structure and measured intelligence are unclear. This is partly because the evidence to-date comes from mostly small and heterogeneous studies. Here, we report brain structure-intelligence associations on a large sample from the UK Biobank study. The overall N = 29,004, with N = 18,426 participants providing both brain MRI and at least one cognitive test, and a complete four-test battery with MRI data available in a minimum N = 7201, depending upon the MRI measure. Participants’ age range was 44–81 years (M = 63.13, SD = 7.48). A general factor of intelligence (g) was derived from four varied cognitive tests, accounting for one third of the variance in the cognitive test scores. The association between (age- and sex- corrected) total brain volume and a latent factor of general intelligence is r = 0.276, 95% C.I. = [0.252, 0.300]. A model that incorporated multiple global measures of grey and white matter macro- and microstructure accounted for more than double the g variance in older participants compared to those in middle-age (13.6% and 5. 4%, respectively). There were no sex differences in the magnitude of associations between g and total brain volume or other global aspects of brain structure. The largest brain regional correlates of g were volumes of the insula, frontal, anterior/superior and medial temporal, posterior and paracingulate, lateral occipital cortices, thalamic volume, and the white matter microstructure of thalamic and association fibres, and of the forceps minor. Many of these regions exhibited unique contributions to intelligence, and showed highly stable out of sample prediction.
Naturally, this is the same thing that Marxist liars like Steven J. Gould have been denying for decades. So not too surprising, scientifically speaking, and in line with Sesardic’s Conjecture.
(Thanks to Inquisitive Bird for this.) I don’t know of any mass replication studies in neuroscience akin to the reproducibility project for psychology, but there is another way to approach this:
-
Kong, X. Z., Postema, M. C., Guadalupe, T., de Kovel, C., Boedhoe, P. S., Hoogman, M., … & Medland, S. E. (2020). Mapping brain asymmetry in health and disease through the ENIGMA consortium. Human Brain Mapping.
The problem of poor reproducibility of scientific findings has received much attention over recent years, in a variety of fields including psychology and neuroscience. The problem has been partly attributed to publication bias and unwanted practices such as p‐hacking. Low statistical power in individual studies is also understood to be an important factor. In a recent multisite collaborative study, we mapped brain anatomical left–right asymmetries for regional measures of surface area and cortical thickness, in 99 MRI datasets from around the world, for a total of over 17,000 participants. In the present study, we revisited these hemispheric effects from the perspective of reproducibility. Within each dataset, we considered that an effect had been reproduced when it matched the meta‐analytic effect from the 98 other datasets, in terms of effect direction and significance threshold. In this sense, the results within each dataset were viewed as coming from separate studies in an “ideal publishing environment,” that is, free from selective reporting and p hacking. We found an average reproducibility rate of 63.2% (SD = 22.9%, min = 22.2%, max = 97.0%). As expected, reproducibility was higher for larger effects and in larger datasets. Reproducibility was not obviously related to the age of participants, scanner field strength, FreeSurfer software version, cortical regional measurement reliability, or regional size. These findings constitute an empirical illustration of reproducibility in the absence of publication bias or p hacking, when assessing realistic biological effects in heterogeneous neuroscience data, and given typically‐used sample sizes.
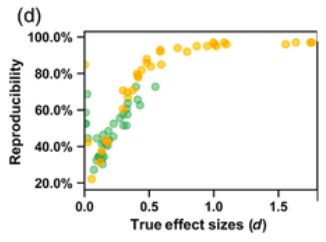
My take away is: do not trust findings from small samples, no matter what statistics the authors report, and especially when they report marginal findings. In general, 3rd parties often can’t even produce the same findings even when they have the same dataset, sometimes called analytic replication, or reproducibility.
Anon adds
Anon writes me and adds:
Real hardcore neuroscience usually involves in vivo/ex vivo human or animal brain tissue. They do very impressive stuff with e.g. how cells connect or how oscillations are generated, and the methodology is very advanced. “Blobology” MR is frankly just not mainstream anymore but this invasive neuroscience has huge respect and I rarely see anybody question its methodology. But when it comes to real-life applications they go beyond the data like the worst old school social psychologists (or the people citing them do).My favorite example is this study from my field: https://www.sciencedirect.com/science/article/pii/S0896627317305494 It is of course very specific but shows many of the typical problems of the field. This is about a sleep-related EEG oscillation (sleep spindles) which are supposed to play a role in memory formation even in humans. There is a literature of small, high RDF [researcher degree of freedom] human studies showing that if you have more of this oscillation you have better overnight memory consolidation, but the problems with that are very much like what is discussed in Button et al. (There is a large negative study which is less frequently cited: https://pubmed.ncbi.nlm.nih.gov/25325488). This is what they did in the current study:– They took <10 mice per experimental condition (who knows how many they had in total? Very easy to hide data like this!). These were transgenic mice – they expressed a channel rhodopsin in a thalamic nucleus so that their interneurons fired when they got a laser stimulation, triggering the oscillation in question. Like I said, the methodology is really high-tech. In another condition they also tested a reversed condition using knockout mice.– Mice learned to be afraid of a tone because they were shocked when they heard it. They slept and they were either 1) not stimulated with the laser 2) stimulated when another oscillation was present 3) stimulated at random (these are the 3 experimental conditions). The question is: when they later hear the tone, do they freeze more frequently (a sign of successful fear learning) if they received the stimulation when this other supposedly crucial oscillation was present?– Yes they do (Figure 1E middle). But! 1) This was a between-subject design, there were different mice in each condition, the same mouse was not tried in all 3. Are we really sure there was nothing like e.g. the non-stimulated mice were just less fearful? Again, we are talking <10 mice per group, and even before anything happened to them there was a strong trend towards this behavior (Figure 1E left). 2) They only froze more often when they heard the tone in exactly the same context (the same type of container)! When they heard the tone elsewhere (Figure 1E right) there was no difference at all. This is literally the opposite of what the title and the abstract claims, that “memory formation” takes place in the presence of these oscillations – only contextual fear is enhanced, that is if you believe the stats. Others also cite this study as strong evidence that this oscillation plays a strong role in strengthening memories in general, obviously even in humans.This is not just a random paper, it was published in hyper-exclusive Neuron with almost 200 citations in 3 years. Most new papers on spindle function (a fringe field but neuroscience is just a bunch of fringe fields) cite it uncritically based on the extraordinary claims in the title and abstract. There are also many like this in other subfields, with the same usual flaws: 1) very high-tech design, but small animal or tissue samples 2) a lot of readily exploited RDF with statistics 3) claims of describing complex psychological functions when what they really investigate is very limited models in animals (“freezing after a conditioned tone/shock pair in rats” as a model of “memory in humans”). I think that fMRI studies are actually much better because there is a will and a way to increase sample sizes, standardize methods and go towards a GWAS-like replicable statistical pipeline. I see little such improvement in this other neuroscience.
