See prior post on self vs. other measurements of personality for a similar take.
Height is the favorite trait in ‘behavioral’ genetics: it’s easy, fast and cheap to measure, continuous as opposed to dichotomous or discrete, highly polygenic, highly heritable, and no one complains to the PC police about it. So we have tons of data about it from twins and other people. There is one particular mega-analysis of twins worth noting:
-
Jelenkovic, A., Hur, Y. M., Sund, R., Yokoyama, Y., Siribaddana, S. H., Hotopf, M., … & Pang, Z. (2016). Genetic and environmental influences on adult human height across birth cohorts from 1886 to 1994. Elife, 5, e20320.
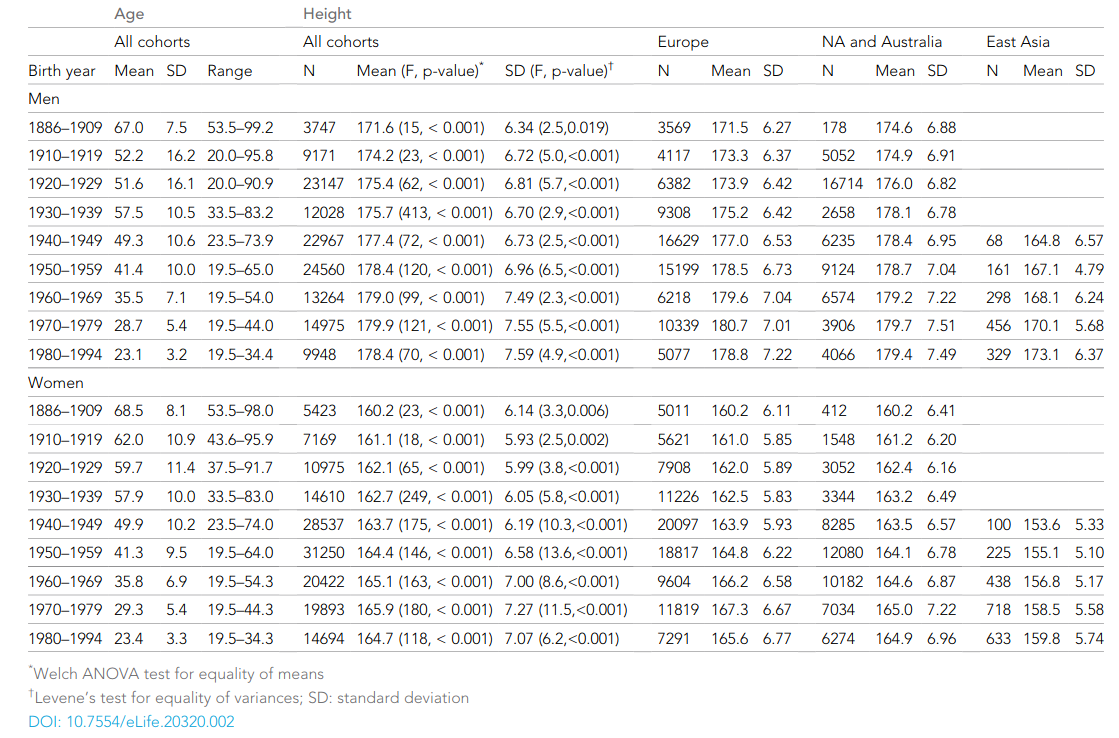
Human height variation is determined by genetic and environmental factors, but it remains unclear whether their influences differ across birth-year cohorts. We conducted an individual-based pooled analysis of 40 twin cohorts including 143,390 complete twin pairs born 1886–1994. Although genetic variance showed a generally increasing trend across the birth-year cohorts, heritability estimates (0.69-0.84 in men and 0.53-0.78 in women) did not present any clear pattern of secular changes. Comparing geographic-cultural regions (Europe, North America and Australia, and East Asia), total height variance was greatest in North America and Australia and lowest in East Asia, but no clear pattern in the heritability estimates across the birth-year cohorts emerged. Our findings do not support the hypothesis that heritability of height is lower in populations with low living standards than in affluent populations, nor that heritability of height will increase within a population as living standards improve.
Besides being absolutely massive at 143k pairs, it is also notable for the lack of a historical trend in heritabilities:

According to the usual Scarr-Rowe reasoning, the worse environments of the past should affect traits like height via stunting and so on, and this environmental variance should somewhat family-linked, thus resulting in larger c² estimates in the past and lower h² ones. Except that we don’t see this above. Instead, it seems to be roughly constant. This does not match the results from certain other traits, mainly educational attainment. (The intelligence heritability is probably also pretty constant, since the oldest studies find quite high values too, but this is not well studied.)
While that is interesting and puzzling in itself, I want to highlight the unusual finding in this meta-analysis: namely lots of visible c² effects, even in recent decades like the 1970s (‘recent’ = 4-5 decade ago!). It beggars belief that family environment really should affect height then, either by parental or sibling effects. Essentially no one was starving or stunted in these cohorts, so there shouldn’t be any such c² variance. So what gives? There are three sources of bias in these estimates. As the authors note:
Important advantages of individual-based data are improved opportunities for statistical modeling and lack of publication bias.This type of analysis is difficult to perform by using literature-based meta-analyses because most of the published studies do not provide the needed statistics by birth-year cohorts. However, our study also has limitations. Countries and/or ethnic-cultural regions are not equally represented and the database is heavily weighted toward populations following Westernized lifestyles. In the classical twin design, parental phenotypic assortment increases dizygotic correlations and thus inflates the shared environmental component when not accounted for in the modeling. In our database, we do not have information on parental height and thus could not take into account assortative mating, which may thus explain part of the shared environmental variation. In addition, most of the height measures were self-reported (Silventoinen et al., 2015), which may bias our analyses toward higher estimates of unique environmental effects due to increased measurement error. However, these sources of bias are unlikely to explain our main result, i.e., relatively similar heritability estimates of adult height over birth cohorts. Finally, since we previously showed that there was no zygosity difference in height variance (Jelenkovic et al., 2015), variance components estimates should not be affected by changes in the proportion of MZ to DZ twins across birth-year cohorts.
So, there are three problems, two mentioned above (unmodeled assortative mating, and self-report data). The final one is that zygosity is often based on survey data, not DNA data. There will be a small error rate, meaning that some people classified as MZ are DZ and some as DZ are MZ. This means that the genetic intraclass correlation for MZs is not 1.00 but some lower value (ignoring somatic mutations), and the value for DZs is more than .50. This will cause the a² estimate to be too low and the c² too high. Thus, the effect of this small bias is the same as that for assortative mating. Self-report data may seem random, but this depends on the kind of error that self-report induces. If it is entirely random error, then this causes both a² and c² to be deflated and e² inflated. However, if there is some systematic bias too that itself has a² or c² causes (which virtually certain, found for personality), then there will be some additional estimation bias of unknown direction. There are some studies that have examined this kind of thing (prior tweet):
-
Elks, C. E., Den Hoed, M., Zhao, J. H., Sharp, S. J., Wareham, N. J., Loos, R. J., & Ong, K. K. (2012). Variability in the heritability of body mass index: a systematic review and meta-regression. Frontiers in endocrinology, 3, 29.
Evidence for a major role of genetic factors in the determination of body mass index (BMI) comes from studies of related individuals. Despite consistent evidence for a heritable component of BMI, estimates of BMI heritability vary widely between studies and the reasons for this remain unclear. While some variation is natural due to differences between populations and settings, study design factors may also explain some of the heterogeneity. We performed a systematic review that identified 88 independent estimates of BMI heritability from twin studies (total 140,525 twins) and 27 estimates from family studies (42,968 family members). BMI heritability estimates from twin studies ranged from 0.47 to 0.90 (5th/50th/95th centiles: 0.58/0.75/0.87) and were generally higher than those from family studies (range: 0.24–0.81; 5th/50th/95th centiles: 0.25/0.46/0.68). Meta-regression of the results from twin studies showed that BMI heritability estimates were 0.07 (P = 0.001) higher in children than in adults; estimates increased with mean age among childhood studies (+0.012/year, P = 0.002), but decreased with mean age in adult studies (−0.002/year, P = 0.002). Heritability estimates derived from AE twin models (which assume no contribution of shared environment) were 0.12 higher than those from ACE models (P < 0.001), whilst lower estimates were associated with self reported versus DNA-based determination of zygosity (−0.04, P = 0.02), and with self reported versus measured BMI (−0.05, P = 0.03). Although the observed differences in heritability according to aspects of study design are relatively small, together, the above factors explained 47% of the heterogeneity in estimates of BMI heritability from twin studies. In summary, while some variation in BMI heritability is expected due to population-level differences, study design factors explained nearly half the heterogeneity reported in twin studies. The genetic contribution to BMI appears to vary with age and may have a greater influence during childhood than adult life.
The p values here are very dodgy, but the effects will be there. Quantitatively, they look like this:

So what their meta-regression shows is that DNA-based zygosity measurement results in 4%point higher a² estimate, and self-report results in 4.8%points lower a² estimate. Together, this is quite sizable 8.8%points. To repeat, these values are quite uncertain and may be smaller or larger (probably smaller). This study is about BMI, not height, but height is a component (BMI = weight[kg]/height[m]²). There is another more direct study:
-
Macgregor, S., Cornes, B. K., Martin, N. G., & Visscher, P. M. (2006). Bias, precision and heritability of self-reported and clinically measured height in Australian twins. Human genetics, 120(4), 571-580.
Many studies of quantitative and disease traits in human genetics rely upon self-reported measures. Such measures are based on questionnaires or interviews and are often cheaper and more readily available than alternatives. However, the precision and potential bias cannot usually be assessed. Here we report a detailed quantitative genetic analysis of stature. We characterise the degree of measurement error by utilising a large sample of Australian twin pairs (857 MZ, 815 DZ) with both clinical and self-reported measures of height. Self-report height measurements are shown to be more variable than clinical measures. This has led to lowered estimates of heritability in many previous studies of stature. In our twin sample the heritability estimate for clinical height exceeded 90%. Repeated measures analysis shows that 2–3 times as many self-report measures are required to recover heritability estimates similar to those obtained from clinical measures. Bivariate genetic repeated measures analysis of self-report and clinical height measures showed an additive genetic correlation >0.98. We show that the accuracy of self-report height is upwardly biased in older individuals and in individuals of short stature. By comparing clinical and self-report measures we also showed that there was a genetic component to females systematically reporting their height incorrectly; this phenomenon appeared to not be present in males. The results from the measurement error analysis were subsequently used to assess the effects of error on the power to detect linkage in a genome scan. Moderate reduction in error (through the use of accurate clinical or multiple self-report measures) increased the effective sample size by 22%; elimination of measurement error led to increases in effective sample size of 41%.
So in general, beware of small effects since they can result from one or a combination of various of smaller biases. Larger effects are less likely to be the result of some bias instead of a signal.