While we don’t take anecdotes too seriously on this blog, here I will present a small one. If you are one of those who have purchased genetic test results from consumer genomics companies such as 23andme or ancestry.com, you can download your ‘raw’ data and upload it elsewhere (or look at it yourself). I saw ‘raw’ data because these data are not by any means the raw, which would be cell files (uninterpretable gibberish to 99.9% of people) but they give you the genotype calls: a list of which 2 variants you have for each tested location. It looks like this (23andme):
# This data file generated by 23andMe at: Mon Mar 3 11:48:42 2014 # # Below is a text version of your data. Fields are TAB-separated # Each line corresponds to a single SNP. For each SNP, we provide its identifier # (an rsid or an internal id), its location on the reference human genome, and the # genotype call oriented with respect to the plus strand on the human reference sequence. # We are using reference human assembly build 37 (also known as Annotation Release 104). # Note that it is possible that data downloaded at different times may be different due to ongoing # improvements in our ability to call genotypes. More information about these changes can be found at: # https://www.23andme.com/you/download/revisions/ # # More information on reference human assembly build 37 (aka Annotation Release 104): # http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9606 # rsid chromosome position genotype rs4477212 1 82154 AA rs3094315 1 752566 AA rs3131972 1 752721 GG rs12124819 1 776546 AG rs11240777 1 798959 GG rs6681049 1 800007 CC rs4970383 1 838555 CC rs4475691 1 846808 CC rs7537756 1 854250 AA rs13302982 1 861808 GG rs1110052 1 873558 GT rs2272756 1 882033 AG rs3748597 1 888659 CC
My 23andme genome has been public since 2012, so I am not disclosing anything in particular here. I think genomes will be mostly public in the future, so there’s little harm in publishing them now (unless you plan to become a serial killer). So, a bit of family history. I have psoriasis, a skin disorder that essentially makes you look sun burned semi-permanently at certain spots on the body. It’s a genetic disease with a high heritability of 68% in a sample of 10k twins. My mother also has the disease and my little brother has it but only weakly so. My dad does not have it, nor my mother’s mother. I have all of their genomes and uploaded them to impute.me, which will give the genetic predictions. They look like this:
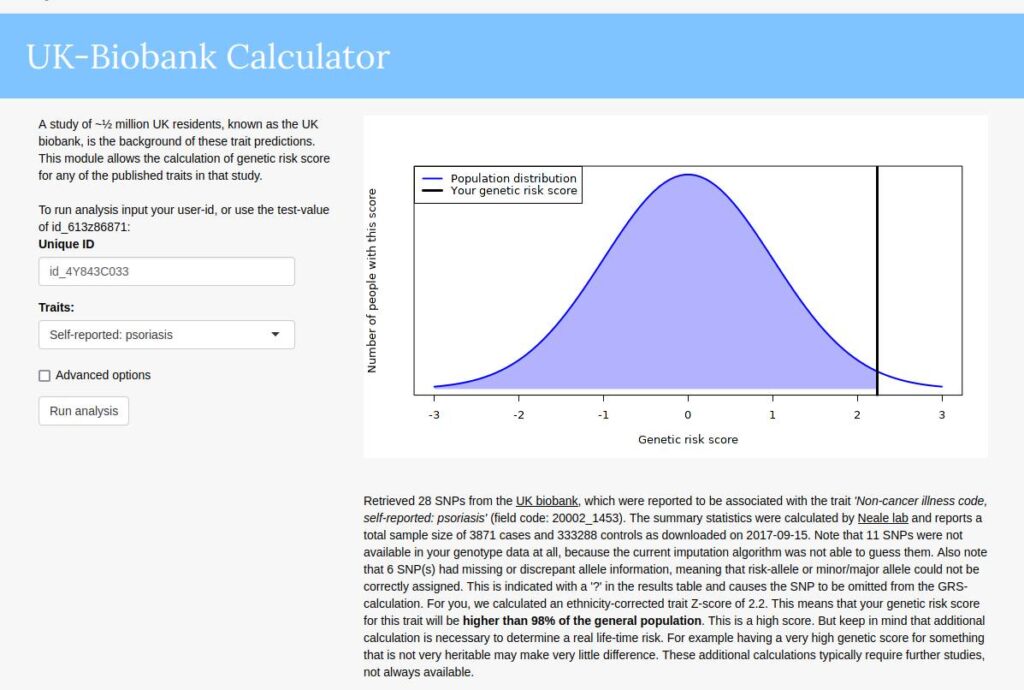
So I get 98% centile on their model based on UKBB GWAS. Rest of family:
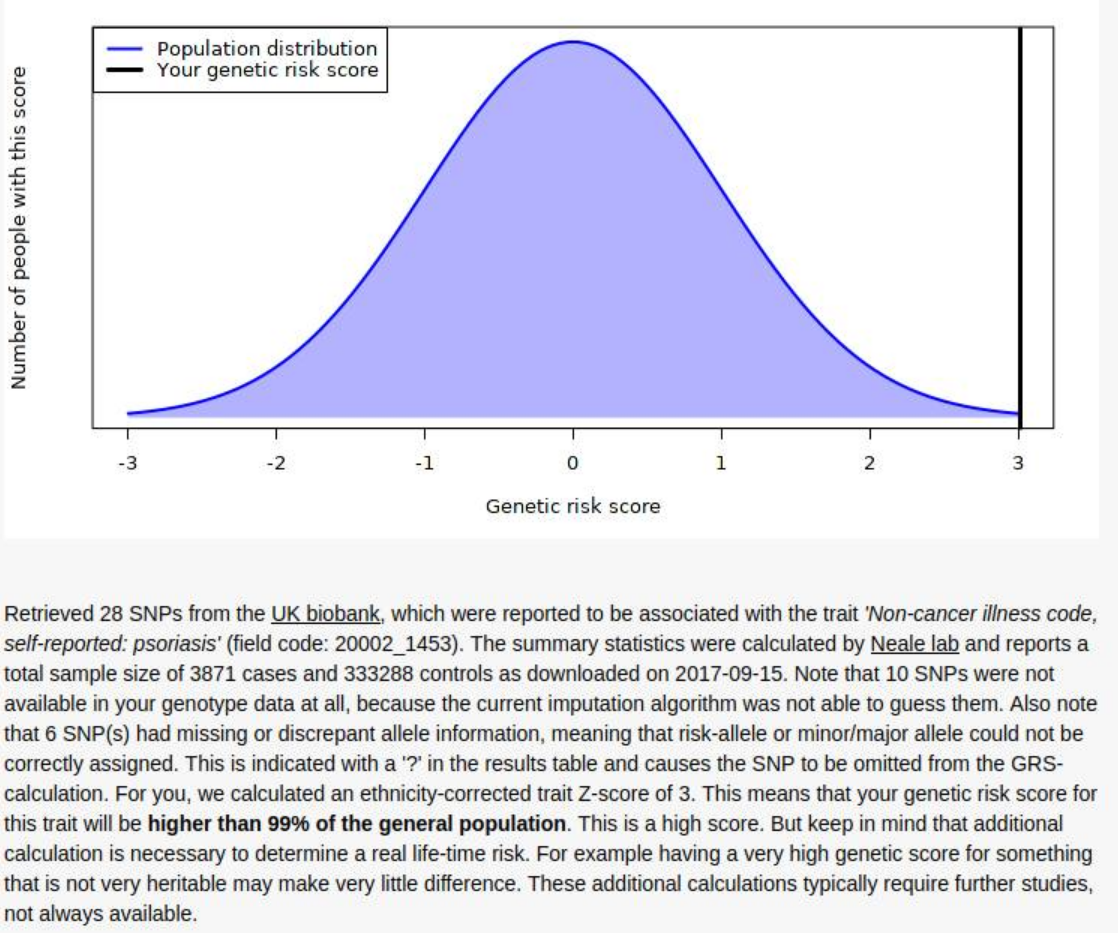
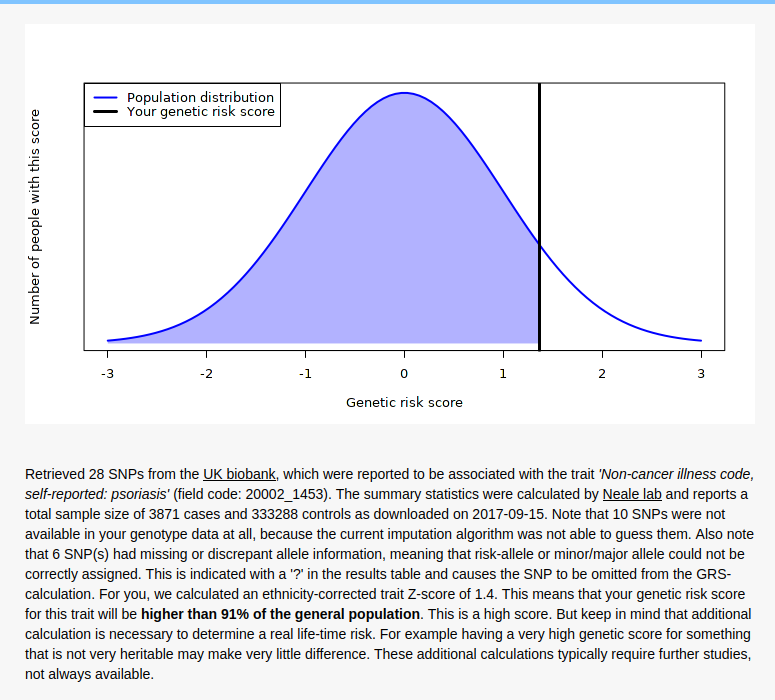

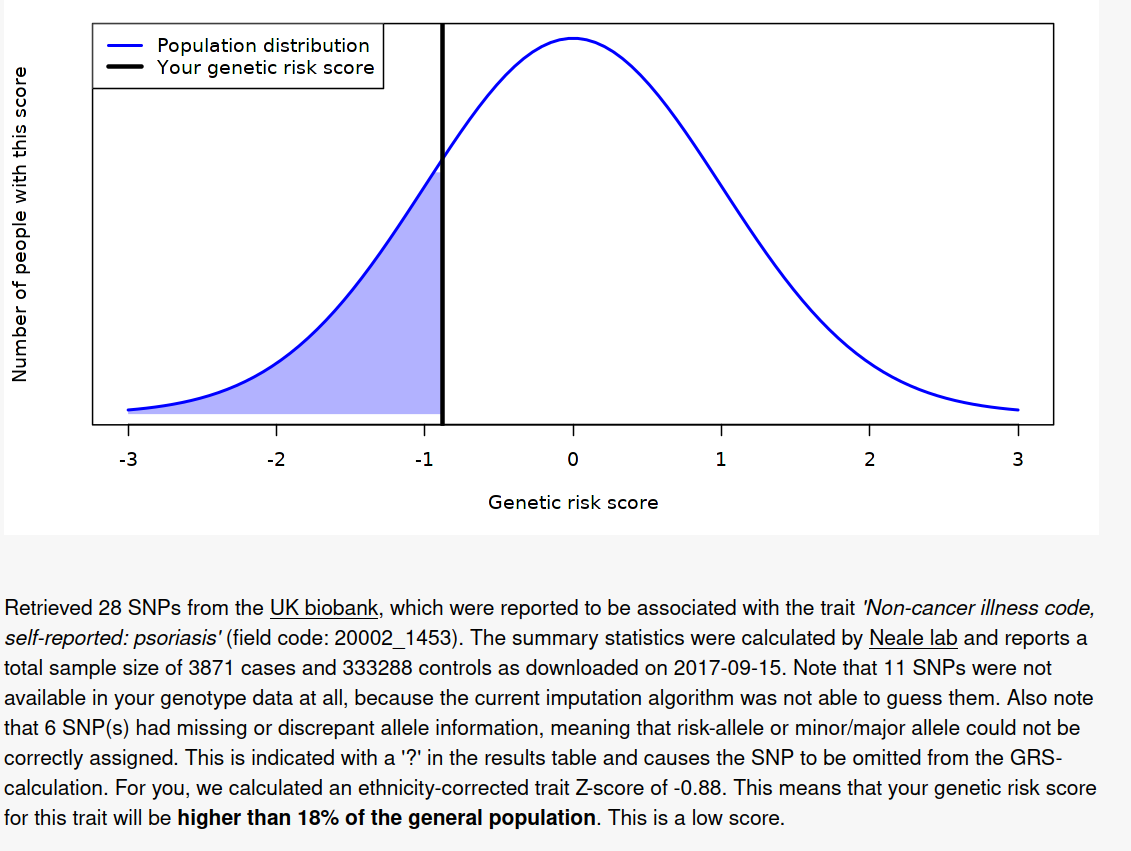
So the results are surprisingly 100% congruent with the phenotype. It’s not clear how good their model is. The model itself can be found here, but no accuracy statistics are given for it, just the list of SNP results (1.8 GB TSV!). I found a much smaller 2011 GWAS (2815 case-control samples and 858 family samples) which had AUC = .72, and a 2017 bigger one (“combined effective sample size >39,000 individuals”) with AUC = .76, so the UKBB model (3871 cases and 333288 controls) will be quite a bit better, maybe AUC = .85.